
Noncanonical translation initiation mechanisms
Viral and cellular RNAs can bypass some or all of the above steps of translation initiation using alternative pathways mediated by highly structured RNA elements in the 5′-UTR. We have studied translation initiation on the model cricket paralysis virus intergenic region internal ribosome entry site (IRES) and the hepatitis C virus IRES to understand how 80S ribosome assembly is achieved by subverting the standard initiation pathway during host infection. Our current research in this area is focused on using biochemical and structural methods to examine repeat-associated non-AUG (RAN) translation. This will help us understand fundamental aspects of scanning, start codon selection, and 80S ribosome assembly and how these steps are mis-regulated in human repeat expansion diseases.

Figure 7
Figure 7 The Hepatitis C virus internal ribosome entry site bypasses the scanning process by binding directly to 40S subunits, leading to initiation without many factors. Structure of human 40S-IRES complex showing IRES RNA in blue, 40S subunit in yellow, and RACK1 protein in yellow. We have long studied structure-function relations using viral IRES.
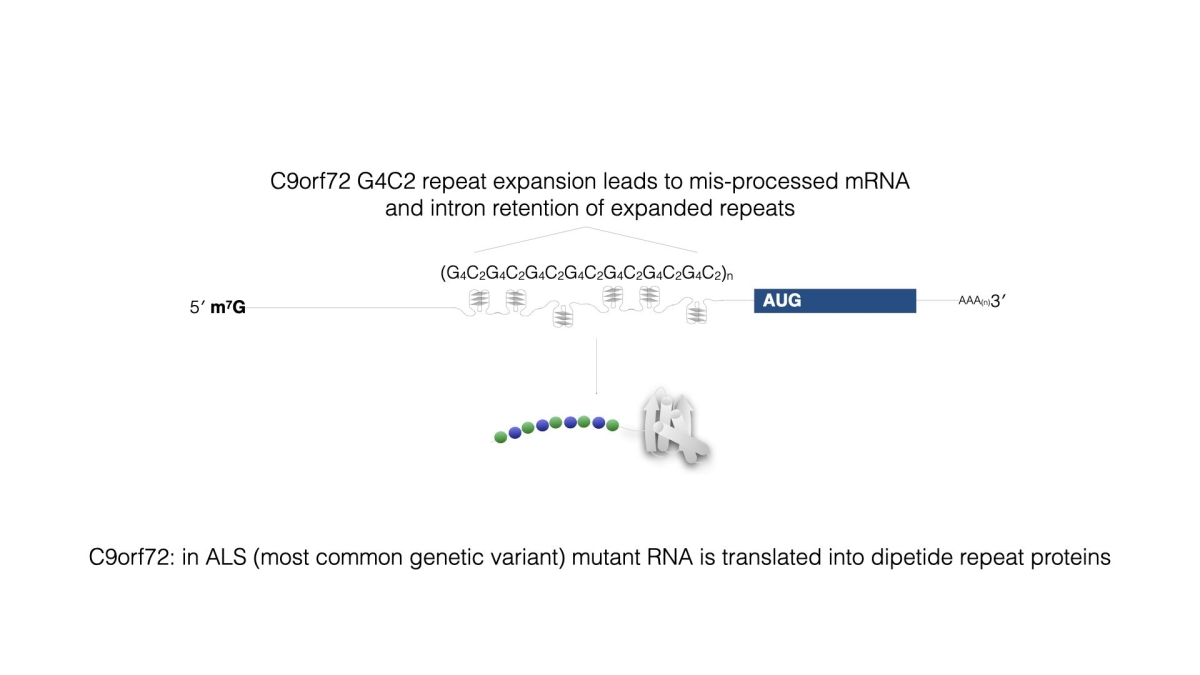
Figure 8
Figure 8 Aberrant initiation and translation can lead to human disease. Repeat expansions in the human C9orf72 gene leads to incorrect splicing and translation of the repeat-containing RNA. The 5’ UTR does not contain a cognate initiation codon, but initiation occurs a CUG, likely influenced by the presence of G-rich structures in the mRNA, and translation creates aggregation-prone dipeptide repeat proteins.
